Using R
This instructional guide will provide you with many ways on how to use R on the CQUni HPC system.
R is a popular programming language which is commonly used for statistical computing and graphics. R can be used in different stages of research such as data visualisation, cleaning and analysis.
To run R programs on CQUniversity’s High Performance Computing system, we can use a graphical interactive development environment (IDE), such RStudio, or via the command line using R directly. We can also submit R jobs to the HPC scheduler to run many R programs non-interactively
To use the R software on the HPC, you will need the following:
- Access to the HPC system (Contact HPC support if you need an account created).
- A connection to the HPC System, see Graphical connection via Open on Demand for information on how to do this.
For those who already have a HPC account and are using a “graphical” connection, you should be able to start RStudio by issuing the following command (inside a terminal session ):
module load rstudio/2024.12.1rstudio
Note, if you get any issues with starting rstudio, try using the following command instead:
rstudio --disable-gpu
What versions of R are available on the HPC system?
It should be noted that it is most likely that the version of R you wish to use is not the default version available when you first log on to CQUniversity’s High Performance Computing facility.
You can use the software module command module avail, to highlight all of the HPC software that is available to load.
It is important to load your R software module you wish to use each time to open a new HPC session, command prompt/terminal as well as have it included in your HPC submission scripts.
A subsection of R software modules that are available to load include:
RR/4.3.2-gfbf-2023a
R/4.4.2-gfbf-2024arstudio
rstudio/2024.12.1
Running and editing R code via a graphical interactive development environment (IDE)
- Connect to the CQUniversity HPC system through using graphical connection, instruction on how to do this can be found here.
- To launch the R IDE RStudio, you will need to do the following:
- Launch the ‘Mate Terminal’ located on your desktop
- Ensure the R version you wish to use is loaded.
3. Ensure the R version you wish to use is loaded.
You can check for any currently loaded R modules using:
which rstudio
/apps/software/rstudio/1.2.1335/bin/rstudio
Transferring files to and from the HPC system
Before you can run your R scripts, you will most likely need to upload your R programs and data from your computer to the HPC system. Instructions on how to do this can be found here.
Once you have uploaded your programs and data, you can then run the R code directly on the HPC system using the instructions provided above.
You can also use the same process of uploading your files to then download the results and anything else you need back to your local computer/s.
Sample Coding on RStudio (interactively solving R jobs
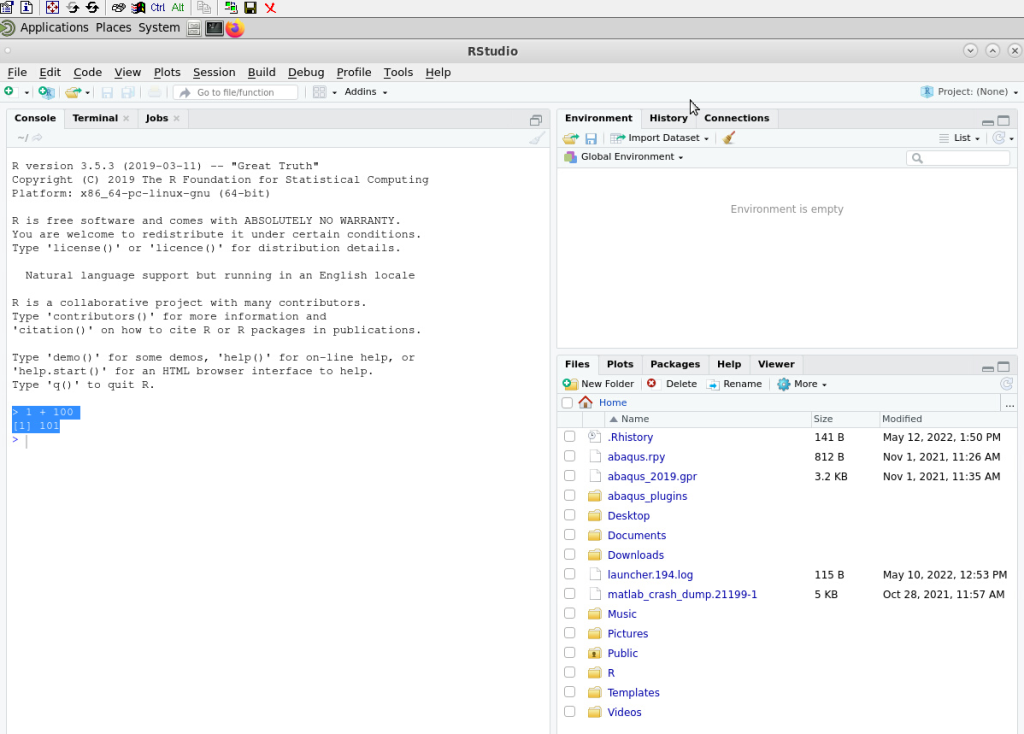
To test if our codes are working in RStudio, we can perform basic mathematical operations such as:
> 1 + 100
That should give us an output:
[1] 101
Solving R jobs non-interactively
One of the benefits of using the HPC system is that you can submit 1 to many jobs to the HPC scheduler. Using the HPC scheduler, you can request more resources (such as CPU’s) which can dramatically improve the processing execution time.
To solve a R job non-interactively, you will need to create a R HPC scheduler script. Instructions of how to do this and some examples can be found at R Sample Scripts.

